Life overwhelms us again and again.
Just imagine, how sweet sugar cane juice is, but it’s genome structure is so complex that it puzzled scientists for years!!!
Sugarcane is grown in over a hundred countries for its sweet juice used in drinks.
But did you know it has many other uses too?
Rum: A popular fermented drink, made from sugarcane.

Bagasse: After extracting the juice, the leftover fibrous material is called bagasse, which is used as a biofuel.
Molasses: This thick syrup is often used to feed animals.
Additionally, sugarcane is used to produce ethanol, another type of biofuel. Ethanol is use in USA, Brazil, China, Canada, Argentina, Thailand and member countries of European union.
In our densely populated world, ethanol takes as a vital asset for its importance as fuel in this worldwide escalating demand.
So more production of sugarcane can solve the demand of biogas. With proper genetic engineering process and DNA sequencing can transform this problem to power for humankind.
Here comes the problem!!
The sugarcane genome presents a tougher puzzle compared to other plants and make it trickier to unlock its full potential.
Why sugarcane genome is so complex? What differs sugarcane’s DNA than others?
I’m sure this question has been bouncing around in your mind.
No worries, let me clear it up for you!
Sugarcane’s genome is so complex because of 2 reasons:-
1.The sugarcane genome is just really big, which makes things a bit more complicated for scientists.
2. It contains more copies of chromosome than a typical plant ( this feature is called polyploidy)
What actually polyploidy means?
Poly means A prefix meaning “Many“
Ploidy means the total number of complete sets of chromosomes in a single cell.
Diploid describes a cell that contains two copies or two sets of each chromosome.
Same as,
monoploid (1 set of chromosome, Example:- Unfertilized eggs of Bees, Ants),
triploid (3 sets-Seedless watermelons and Seedless banana), tetraploid (4 sets- Wheat, Cotton, and Brussel sprouts), pentaploid (5 sets-Betula kenaica), hexaploid (6 sets-kiwifruit), heptaploid or septaploid (7 sets-Siberian Sturgeon) etc.
In one copy of the human genome consists of approximately 3 billion base pairs
and guess what?
Sugarcane has about 10 billion base pairs in their genome!!!!
That’s make the DNA of sugarcane large and complex and so it becomes difficult for the researcher to determine the location of specific gene in their chromosome which is known as gene mapping.
So, finally it become clear in details about polyploidy and complex structure of Sugarcane.
Researchers are poised to unravel this complex puzzle by applying novel approaches that turns this complexity into elegant simplicity.
But HOW?
Researchers untangle this puzzle structure with the help of various gene sequencing techniques, taking in HiFi sequencing (A newly crafted method)
But first, let’s learn some of gene sequencing methods?
Some gene sequencing methods
Long-read sequencing methods
1.Single molecule real time (SMRT) sequencing
2.Nanopore DNA sequencing
Short-read sequencing methods
1.Polony sequencing
2.Illumina (Solexa) sequencing
3.454 pyrosequencing
4.Massively Parallel Signature Sequencing (MPSS)
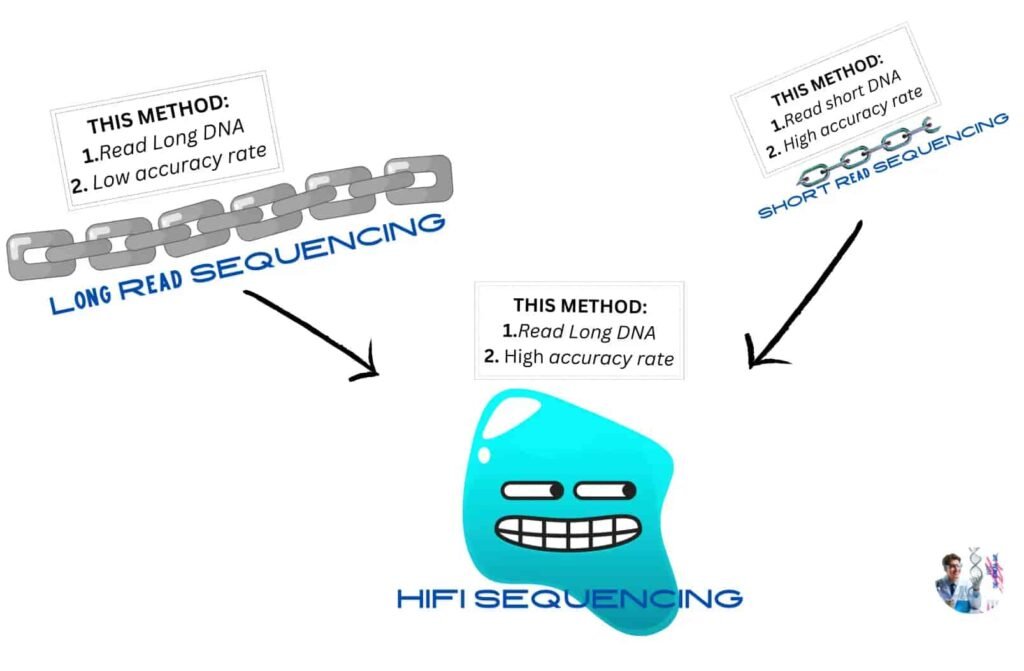
Because of the large size of the DNA of Sugarcane traditional long read sequencing could be used .The target of long read sequencing is to read or understand very long DNA such as whole chromosome. But the problem is the accuracy rate of the method is very low. That means using long-read sequencing, the large DNA of sugarcane can be sequenced faster, but it might still have errors.
Moving to Short-read sequencing methods which aims is to read short fragments of DNA and the good news is the accuracy rate of this method is more higher.
Although it is more accurate method, however it can consume countless time for sequencing the large DNA of sugarcane.
Combining the best parts of short reads and long reads-highly accurate long-read sequencing or HiFi sequencing method appear as a angel for the researchers, enabling to solve complex genome of sugarcane. By using the method it becomes possible not only read or sequence of large-size DNA but also achieve 99.9% accuracy or even more. Including time as well money can be saved easily by using this effective method.
This reference genome (a digital database of nucleic acid sequence) which have gotten by using HiFi sequencing, make it convenient to compare with other crops which are properly studied and this comparison opens ultimate door of knowledge indicating that which genes are highly expressed during sugar production, which genes are significant for the resistance of disease, which genes are helpful to grow sugarcane, sucrose metabolism genes, photosyntheses revoted genes etc.
NOTE: From this study a cultivar of sugarcane, OR510 is used
Faculty investigator at the HudsonAlpha Institute for Biotechnology, Jeremy Schmutz said, “This was the most complicated genome sequence we’ve yet completed, It shows how far we’ve come. This is the kind of thing that 10 years ago people thought was impossible. We’re able to accomplish goals now that we just didn’t think were possible to do in plant genomics.”
First author of the paper, Adam Healey who is also a researcher at HudsonAlpha said, “When we sequenced the genome, we were able to fill a gap in the genetic sequence around brown rust disease. There are hundreds of thousands of genes in the sugarcane genome, but it’s only two genes, working together, that protect the plant from this pathogen. Across plants, there are only a handful of instances that we know of where protection works in a similar way. Better understanding of how this disease resistance works in sugarcane could help protect other crops facing similar pathogens down the road.”
This study involved collaborations with institutes from around the world, including
Australia (CSIRO Agriculture and Food, Queensland Alliance for Agriculture and Food Innovation/ARC Centre of Excellence for Plant Success in Nature and Agriculture — University of Queensland, Sugar Research Australia);
France (CIRAD, UMR-AGAP, ERCANE);
Czech Republic (Institute of Experimental Botany of the Czech Academy of Sciences); and
The United States (Corteva Agriscience, Joint BioEnergy Institute).
The genome was sequenced at the Joint Genome Institute (JGI) with work completed at the JGI partner laboratories, the Arizona Genomics Institute and the HudsonAlpha Institute for Biotechnology.
The work may be challenging, but each step forward brings us closer to a world where sugarcane’s potential is fully realized. So, as we continue to explore this sweet puzzle, one thing is clear: the future of sugarcane is bright, and its story is just beginning.
What do you thing?